Sequencing Choices: 16S or WGS?
- 23rd September 2019
- Posted by: Adele Mcmeekin
- Categories: Articles, Single Cell Analysis

As sequencing becomes more affordable and more options are available, the next big decision is what type of sequencing to use for a research study. In this blog post, we take a short look at two sequencing methods – 16S and whole-genome sequencing (WGS) – and find out the background to each method and its unique characteristics.
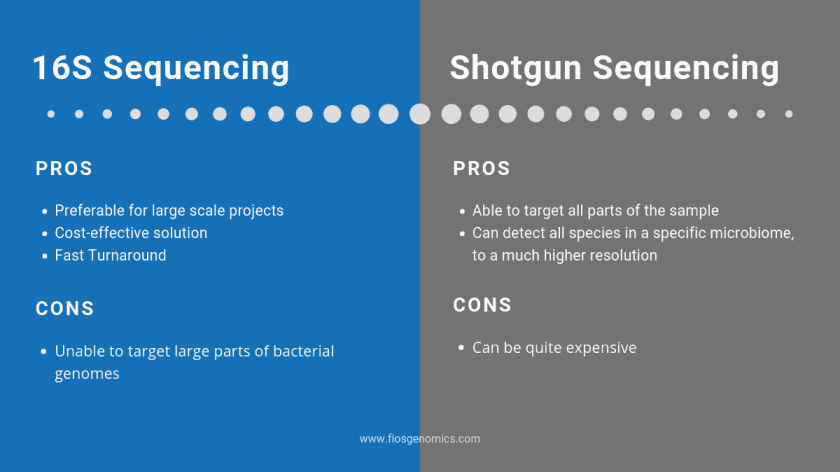
16S Sequencing
16S sequencing specifically looks at the 16S ribosomal RNA (rRNA) gene in bacteria. The utility of 16S was first highlighted in the 1960s by Dubnau et al. who noted conservation of the sequence in Bacillus species. As a highly conserved gene sequence, the 16S rRNA must be a critical component for cellular function.
While the rate of mutation for the 16S genes is not known, and could be different depending on the species, the 16S rRNA gene can still used for identification as the rate of change has a greater impact on relationships of species that are more distantly related. Work by Woese followed as the 16S rRNA sequence became more widely used for bacterial identification and taxonomy.
16S rRNA gene sequences for a large number of strains has been found, meaning that new and unknown sequences are able to be compared against previously sequenced strains. The benefits to 16S are that is relatively cheap and quick to carry out, meaning that it is available for a wide range of research and budgets. The downside however is that it targets a very small portion of the bacterial genome.
While 16S is present in almost all species of bacteria, it isn’t a universal gene. Bacteria could therefore be missing from the results even though they were present. Some bacterial species have multiple 16S genes. With such dynamic genomes, by sequencing one small fragment and assigning a species based on that, information could be lost on the specific strain of bacteria.
Whole-Genome Sequencing
Before WGS became more widely utilised, sequencing used more manual methods which made whole genomes difficult or too time-consuming to collect. After the move into the second and third ‘generations’ of DNA sequencing, whole genomes could be sequenced more easily. Shotgun metagenomics sequences everything that’s present in a sample. This gives a full picture of what species are living in a sample at a much higher resolution. This allows for optics down to strain level, rather than just general family. One disadvantage to shotgun metagenomics is that it can be expensive when compared to 16S sequencing.
New sequencing methods including high-throughput methods use a basic shotgun strategy. This is where longer pieces of DNA are broken down into smaller sections and sequenced randomly. Once sequenced, the sections need to be realigned back into the order of the original DNA unit.
Fios’ Sequencing Analysis Services
At Fios Genomics, we offer a wide range of analysis services. We are platform-agnostic, enabling us to analyse your data no matter the platform you choose for sequencing. Get in touch today to find out how we can help your research.
Leave a Reply
You must be logged in to post a comment.

