Genome Sequencing
- 4th December 2018
- Posted by: Breige McBride
- Category: Articles

Genome sequencing is hugely important because it decodes the sequence of A’s, T’s, G’s, C’s (and sometimes U’s) that make up a genetic code. Once the genome is sequenced and the order of bases realised, work can then begin on deciphering what the strings of letters mean and what they code for.
The first human genome took 13 years to be sequenced. Nowadays, a human genome can be sequenced in a day due to major advances in the technologies available.
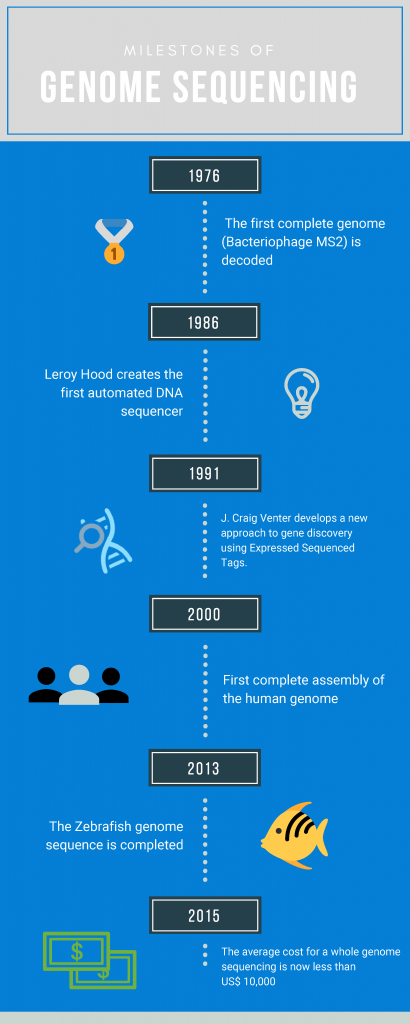
In 1976, the first bacterial genomes were completely sequenced. Bacteriophage MS2 is one of the smallest genomes sequenced, with 3,569 base pairs. In 1996, nearly 20 years later, the first organism had its entire genome sequenced. Haemophilus influenzae, a bacterium, was sequenced and found to have 1,830,140 base pairs of DNA in a singular chromosome. Since then, hundreds of animals and other organisms have had their genomes sequenced.
The largest genome ever sequenced is the Japanese canopy flower Paris japonica. Its genome is 50 times larger than the human genome with 149 billion base pairs encoded. In contrast, the smallest genome of an organism has only 2.25 million base pairs – Encephalitozoon intestinalis is a parasite with a genome 66,000 times smaller than the Japanese flower.
Sequencing a Plant’s Genome vs Animal’s
Plants usually have larger genomes compared to animals. Having a large genome does not correlate with having the most chromosomes, however. The Atlas blue butterfly (Polyommatus atlantica) has roughly 450 chromosomes, over 100 more than the plant species with the most chromosomes (the black mulberry, Morus nigra, which has 308). Plant species are notorious for having multiplication and duplications of their genome.
Polyploidy, the state where a plant has multiple copies of its genome, occurs when chromosomes are incorrectly sorted during cell division. Each gamete should have half the genome of the plant, one copy of each chromosome, making it a haploid cell. A diploid cell (a cell with two copies of each chromosome, making up the entire genome) should not occur during cell division until two gametes have fused together. If two diploid gametes fuse together, a tetraploid offspring (with four sets of chromosomes) is created.
Platypus – Genomic Mixture?
It’s not only plants that have interesting genomes. In animals, the platypus has an interesting mix of both mammalian and reptilian genes. The platypus is a monotreme, one of the three groups of extant mammals. The female platypus produces milk (like other mammals) but also lay eggs, while the males have venomous spurs on their ankles (like reptiles). In studying its genome, similarities between proteins found in reptile and platypus venom were highlighted. The milk protein genes have also been evolutionarily kept, even while eggs are laid by the females and no live births occur. The platypus genome enables deeper analyses to be carried out on other mammalian genomes, whilst also aiding conservation efforts for the species.
Automated Sequencing
The invention of the ‘automated’ sequencer in 1986 helped to speed up genome sequencing. Over 9,000 bases a day could be scanned on the first automated sequencer. Nowadays, over two million bases can be scanned due to the increase in automation and more refined sequencing technologies. The development of Expressed Sequence Tags (ESTs) by J. Craig Venter in 1991 also enabled an increase in the speed of sequencing. ESTs are short sections of complementary DNA sequences, used to identify genes and their locations. Each EST can be mapped to individual chromosomes, allowing for a better understanding of where genes are located in human DNA.
For human genomes, after taking 10 years to get a draft in 2000, and 13 in total for a completed sequence, automated sequencers are now at the stage where, in less than a week, a full genome can be sequenced. Also, the cost of sequencing has dramatically decreased. It is possible nowadays to sequence a whole genome with less than $10.000. New DNA strands and ESTs can be compared against the reference human genome with greater speed and ease – although with only one reference genome for humans, a large amount of genetic diversity is missed out through using a small selection of people to build up the reference.
Your Research
We know that genome sequencing can still be an expensive stage of your research. If you need support choosing the right platform or data type, or if you require analysis, contact Fios today. No matter what organism you’re working with, or the size of your dataset, we are here to help you get the most out of your data.
References:
The largest eukaryotic genome of them all?
Genome analysis of the platypus reveals unique signatures of evolution
Whole-genome random sequencing and assembly of Haemophilus influenzae Rd
Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene
Definition of Eight Mulberry Species in the Genus Morus by Internal Transcribed Spacer-Based Phylogeny
Assembly of a pan-genome from deep sequencing of 910 humans of African descent
Complementary DNA Sequencing: ExpressedSequence Tags and Human Genome Project
Leave a Reply
You must be logged in to post a comment.

